| Next Up Previous Contents |
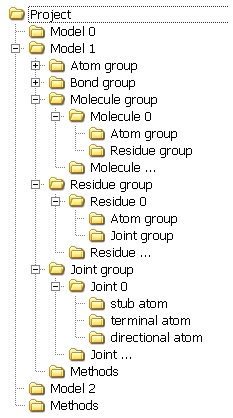
ModelsAll user manipulations are applied to molecular models. In this program, each model corresponds to an individual graphical widow and a file. While multitasking is not supported, actions can be applied either to the active model or to all opened models. For instance, comparison can be applied to all models. A model is a set of atoms and their associations such as bonds, molecules, and residues. Consequently, associations can be included into other associations. For instance, molecules contain atoms and residues, which consequently include atoms and joints. Models can also include operation methods, e.g., simulation options, periodic boundary conditions, etc. Other methods can be common for all models, e.g., geometric measurements. The principal structure of the program can be presented as a tree:
This structure can be seen in models stored in mlm format. The notion of joint deserves specific consideration. A joint defines the connection between residues and consists of three atoms:
The work of the Sequence editor is based on manipulations with joints. Accordingly, only fragments defined in the mlm format can be used to construct chain molecules. |
| Next Up Previous Contents |
| Copyright © 2006-2008 Agile Molecule. All rights reserved |